Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Aguirre, C., Brink, T.T., Guichou, J.F., Cala, O., Krimm, I.(2014) PLoS One 9: e102300-e102300
- PubMed: 25025339
- DOI: https://doi.org/10.1371/journal.pone.0102300
- Primary Citation of Related Structures:
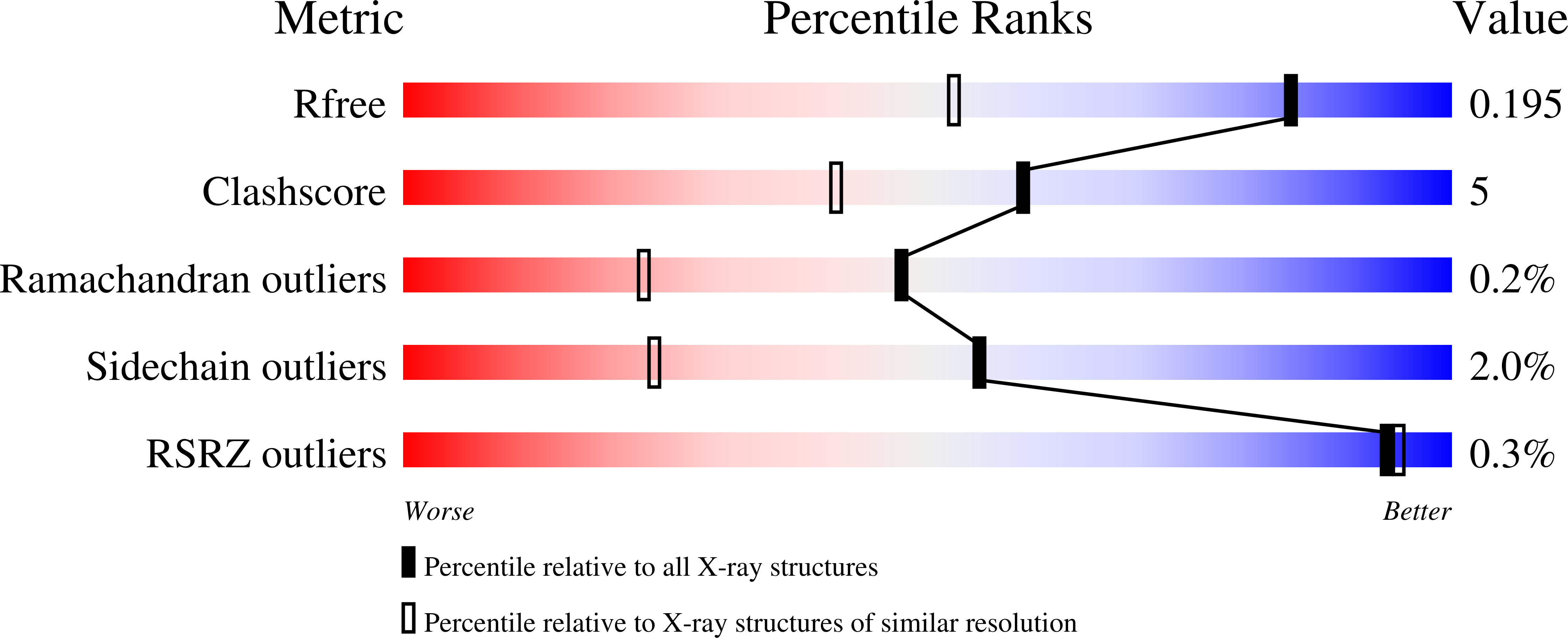
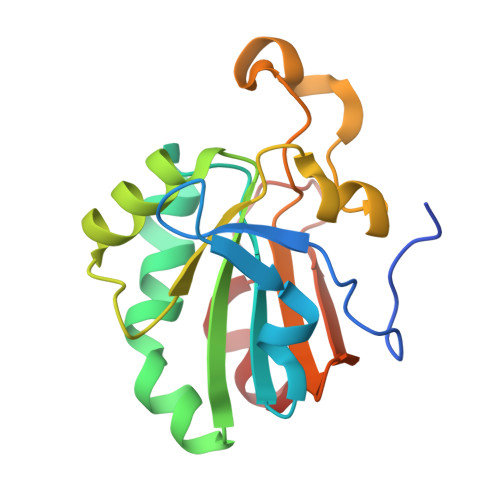
4K7I, 4K7N, 4K7O, 4MMM - PubMed Abstract:
Fragment-based drug design is one of the most promising approaches for discovering novel and potent inhibitors against therapeutic targets. The first step of the process consists of identifying fragments that bind the protein target. The determination of the fragment binding mode plays a major role in the selection of the fragment hits that will be processed into drug-like compounds. Comparing the binding modes of analogous fragments is a critical task, not only to identify specific interactions between the protein target and the fragment, but also to verify whether the binding mode is conserved or differs according to the fragment modification. While X-ray crystallography is the technique of choice, NMR methods are helpful when this fails. We show here how the ligand-observed saturation transfer difference (STD) experiment and the protein-observed 15N-HSQC experiment, two popular NMR screening experiments, can be used to compare the binding modes of analogous fragments. We discuss the application and limitations of these approaches based on STD-epitope mapping, chemical shift perturbation (CSP) calculation and comparative CSP sign analysis, using the human peroxiredoxin 5 as a protein model.
Organizational Affiliation:
Institut des Sciences Analytiques, CNRS UMR 5280, Université de Lyon, Villeurbanne, France.